swc_specification
Information about the SWC file specification. A video describing the specification can be found here: https://vimeo.com/853224668?share=copy
Specification of standard SWC - adapted from (3)
An SWC file (S.W.C. encodes for the last names of its initial designers Ed Stockley, Howard Wheal, and Robert Cannon) is a text file that starts with an optional header section in which each line starts with the symbol #. Some SWC-variants use this section to store information about the data in an orderly fashion, others treat is as a free-text field.
Below the header section, a points matrix with 7 columns follows. It contains the points traced along the neuronal tree. The seven numbers in each row are separated by spaces, and have the following meaning:
Column index |
Field |
Description |
|---|---|---|
1 |
Index |
Sample identifier. A sequential positive integer. |
2 |
Type |
Type identifier. A positive integer. |
3 |
X |
X-position in micrometers |
4 |
Y |
Y-position in micrometers |
5 |
Z |
Z-position in micrometers |
6 |
R |
Radius in micrometers (half the node thickness) |
7 |
Parent |
Parent sample identifier. |
The basic set of types used in NeuroMorpho.org SWC files are:
Type |
ID |
Description |
|---|---|---|
undefined |
0 |
Structure type unknown or unspecified |
soma |
1 |
Soma of a neuron |
axon |
2 |
Axon of a neuron |
basal dendrite |
3 |
Basal dendrite of a neuron |
apical dendrite |
4 |
Apical dendrite of a neuron |
custom |
5 |
Custom type of cell component |
unspecified neurite |
6 |
An unspecified part of a neuron |
glia processes |
7 |
Glial processes |
custom |
>7 |
Custom type of cell component |
Parent defines how points are connected to each other. In a tree, multiple points can have the same ParentID. The first point in the file must have a ParentID equal to -1, which represents the root point. Parent samples must be defined before they are being referred to. By counting how many points refer to the a given parent, the number of its children can be computed.
Soma representation
The soma in SWC can consist of a single root point or multiple points, the first of which is the root. The single-point representation approximates the soma as a sphere of radius R and centered in X, Y, Z. The multi-point representation approximates the soma as a sequence of nodes akin to a neurite branch. Note that representing the soma as a contour tracing of the soma perimeter or a series of contour tracings approximating the soma surface is not consistent with the SWC standard.
SWC Example - adapted from Ascoli et al, 2001

image
This extremely simplified neuronal structure was obtained by extensive pruning of a dentate gyrus granule cell in (4). The left panel shows the digital SWC representation: every node compartment is described by a row containing: - A label (ID or Index, the same numbers reported next to the branches in the right panel - A tag (T or Type, 1 for soma, 3 for dendrite), - Cartesion positions (X,Y,Z in micrometers), - Radius (R) and - Connectivity (C or Parent, representing the label of parent, -1 indicates no parent)
Syntax of basic SWC reconstruction in EBNF - from Nanda et al, 2018
newline = “:raw-latex:`\n`” ;
letter = “A” | “B” | “C” | “D” | “E” | “F” | “G” | “H” | “I” | “J” | “K” | “L” | “M” | “N” | “O” | “P” | “Q” | “R” | “S” | “T” | “U” | “V” | “W” | “X” | “Y” | “Z” | “a” | “b” | “c” | “d” | “e” | “f” | “g” | “h” | “i” | “j” | “k” | “l” | “m” | “n” | “o” | “p” | “q” | “r” | “s” | “t” | “u” | “v” | “w” | “x” | “y” | “z” ;
digit = “0” | “1” | “2” | “3” | “4” | “5” | “6” | “7” | “8” | “9” ;
symbol = “[” | “]” | “{” | “}” | “(” | “)” | “<” | “>” | “‘” |’”’ | “=” | “|” | “.” | “,” | “;” ;
integer = [+|-] digit{digit} ;
double = integer [“.”] {integer} ;
character = letter | double | symbol | “_” | ” “;
header = “#” character {character} newline [“#” character {character} newline] ;
Index = integer ;
Type = integer ;
X = double ;
Y = double ;
Z = double ;
Radius = double ;
Parent = integer ;
node = Index ” ” Type ” ” X ” ” Y ” ” Z ” ” Radius ” ” Parent ;
neuron_tree = node newline {node newline} ;
SWC = [header] neuron_tree ;
Header fields defined in Cannon et al 1998
The original publication that introduced SWC (4) described the header as containing the following fields, although these are not considered a required component of SWC:
Field |
Description |
|---|---|
ORIGINAL_SOURCE |
File type delivered by digitisation equipment |
CREATURE |
Species from which the cell came |
REGION |
Brain region |
FIELD/LAYER |
Location within region |
TYPE |
Cell type |
CONTRIBUTOR |
Name, initials, organisation, e.g. Turner DA, Duke |
REFERENCE |
Where the data has been published |
RAW |
File name of original data |
EXTRAS |
Files containing further information on this cell |
SOMA_AREA |
Area of soma (in mm2) |
SHRINKAGE_CORRECTION |
x, y and z correction factors |
VERSION_NUMBER |
To identify different versions of the same raw data |
VERSION_DATE |
Date this version was created (yyyy-mm-dd) |
SCALE |
Used internally to record applied shrinkage corrections |
Recommendations for Optional Inclusion of Ancillary Information
Individual researchers may choose to specify additional details in the header or footer of the SWC file, see figure below. In particular, the header is most commonly employed to relay metadata information, as originally proposed. Since many of the same metadata elements are frequently employed across studies, we provide here our suggestions based on the most common annotations in use at NeuroMorpho.Org. Specifically, one or more lines starting with the “#” sign should convey the following information, if known: the name of the author(s), dataset, or lab of origin (#contributor); DOI or full bibliographic citation for document describing data (#reference); the animal species and strain or genotype (#creature), sex (#sex), age (#age), and weight (#weight); the anatomical region of the cell body (#region) and the cell type or identifying features (#class); the experimental group (#condition); the labeling or staining (#label); the slicing direction and thickness (#slicing); the objective type and magnification (#microscopy); the physical units (#coordinate), reference frame (#brainspace), and the tracing software (#original_source). It is important to recognize that the above list cannot capture, nor is it always applicable to, all essential details of each neuroanatomy study. Moreover, to truly standardize metadata it would be necessary to define not only the required fields, but also a set of controlled vocabularies to describe the corresponding details. Recent developments in machine learning can greatly facilitate this process.
A second opportunity for increasing the applicability of SWC files is to utilize the footer to convey information regarding synaptic connectivity. Here we adopt the format recently proposed by the fly electron microscopy community. Accordingly, the synaptic connectivity in the SWC file footer should begin with a “#start synapse” line and finish with an “#end synapse” line. These delimiters are useful to avoid accidentally reading as synapses other cellular information that users might want to include in the footer, such as organelle distributions or temporal branch dynamics. In between, each line should describe a synaptic contact with a “#” sign followed by 9 fields: (i) a unique ID for the detected synapse; (ii-iv) the x, y, and z synapse position in coordinate space; (v) the node in the SWC file closest to the synapse; (vi) a binary assignment with 0 indicating an output synapse and 1 indicating an input synapse; (vii) the neurite structural domain, e.g., axon vs. dendrite, analogous to the second data field (type) in the SWC data file; (viii) a unique identifier for the partnering neuron; and (ix) the putative neurotransmitter. Prior to the first synapse data line, a line (always starting with “#”) should list the meaning of these fields.
The above information could be parsed automatically by suitable connectomics analysis platforms. At the same time, because traditional SWC readers interpret the “#” as a comment indicator, the proposed header and footer formats ensure continuous back-compatibility with legacy software.
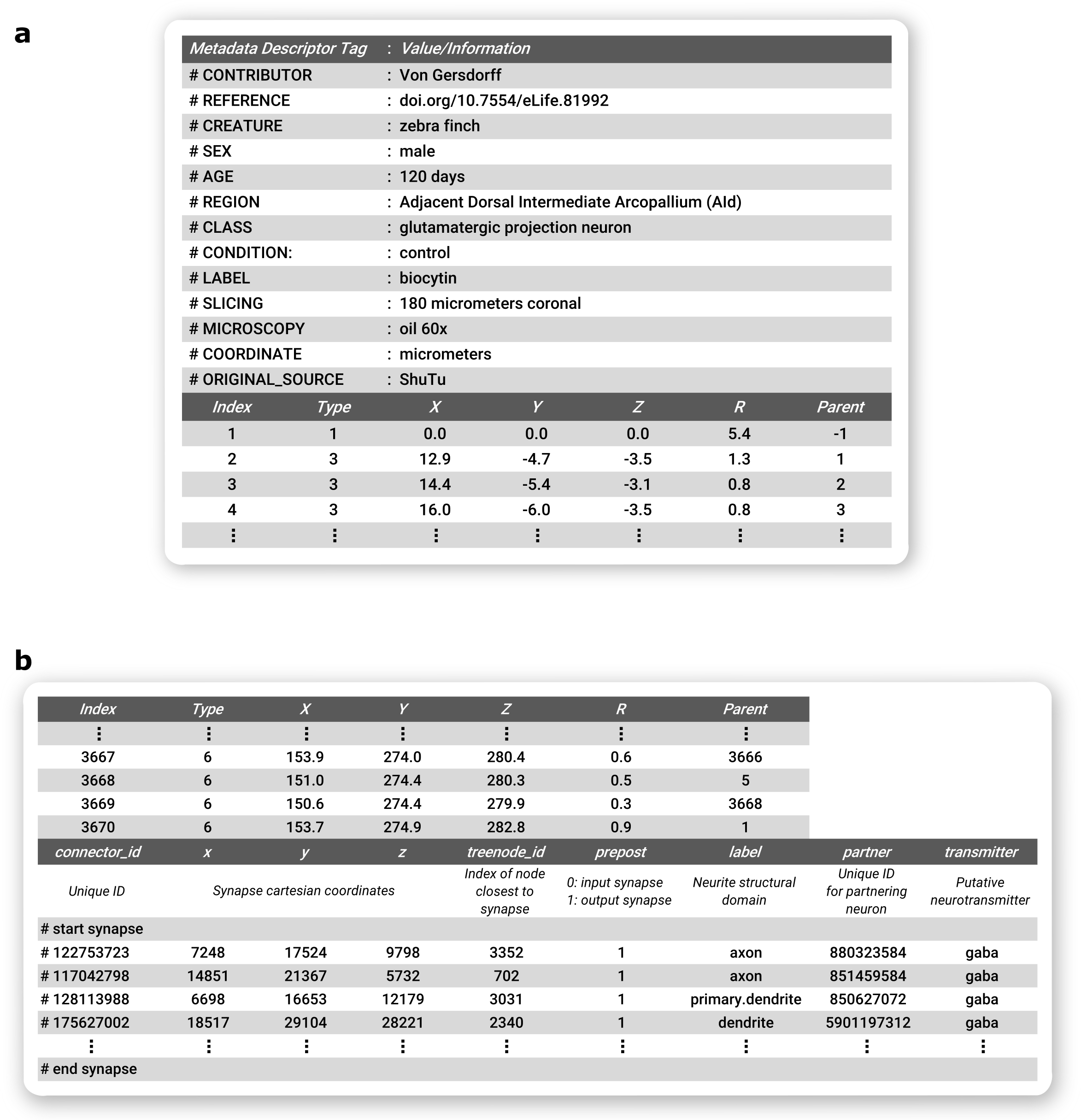
Recommended optional inclusion of ancillary information in SWC Files. a Metadata information included as a header, b Synapse connectivity information in footer.
References
Ascoli GA, Krichmar JL, Nasuto SJ, Senft SL. Generation, description and storage of dendritic morphology data. Philos Trans R Soc Lond B Biol Sci. 2001;356(1412):1131-1145. doi:10.1098/rstb.2001.0905 (PMCID: PMC1088507)
Nanda S, Chen H, Das R, et al. Design and implementation of multi-signal and time-varying neural reconstructions. Sci Data. 2018;5:170207. Published 2018 Jan 23. doi:10.1038/sdata.2017.207 (PMCID: PMC5779069)
https://neuroinformatics.nl/swcPlus/ “SWC plus (SWC+) format specification”
Cannon RC, Turner DA, Pyapali GK, Wheal HV. An on-line archive of reconstructed hippocampal neurons. Journal of Neuroscience Methods. 1998 Oct;84(1-2):49-54. DOI: 10.1016/s0165-0270(98)00091-0. PMID: 9821633.
https://en.wikipedia.org/wiki/Extended_Backus%E2%80%93Naur_form
http://neuromorpho.org/SomaFormat.html Soma format representation in NeuroMorpho.Org as of version 5.3
Contributors
Lydia Ng
Giorgio Ascoli
Bengt Ljungquist
Sumit Nanda